Getting started
To connect to Open OnDemand, visit https://ondemand.cluster.france-bioinformatique.fr
The first page of IFB Open OnDemand will bring you to a login prompt. Use your IFB cluster account to login.
Features
Once you have logged into IFB Open OnDemand, the landing page will display several tabs across the top, including Files, Jobs, Clusters (shell access), Interactive Apps, and My Interactive Sessions. In the center of the page, direct links to popular interactive apps are prominently featured for easy access.

Files
The File menu lets you view and use files in different file spaces: your home directory and the projects directory (/shared/projects). Selecting one of the file spaces takes you to a page that lets you navigate through the space in a graphical frame similar to a file browser on a personal computer. From here you can download, upload, create, delete and open files.

Tip
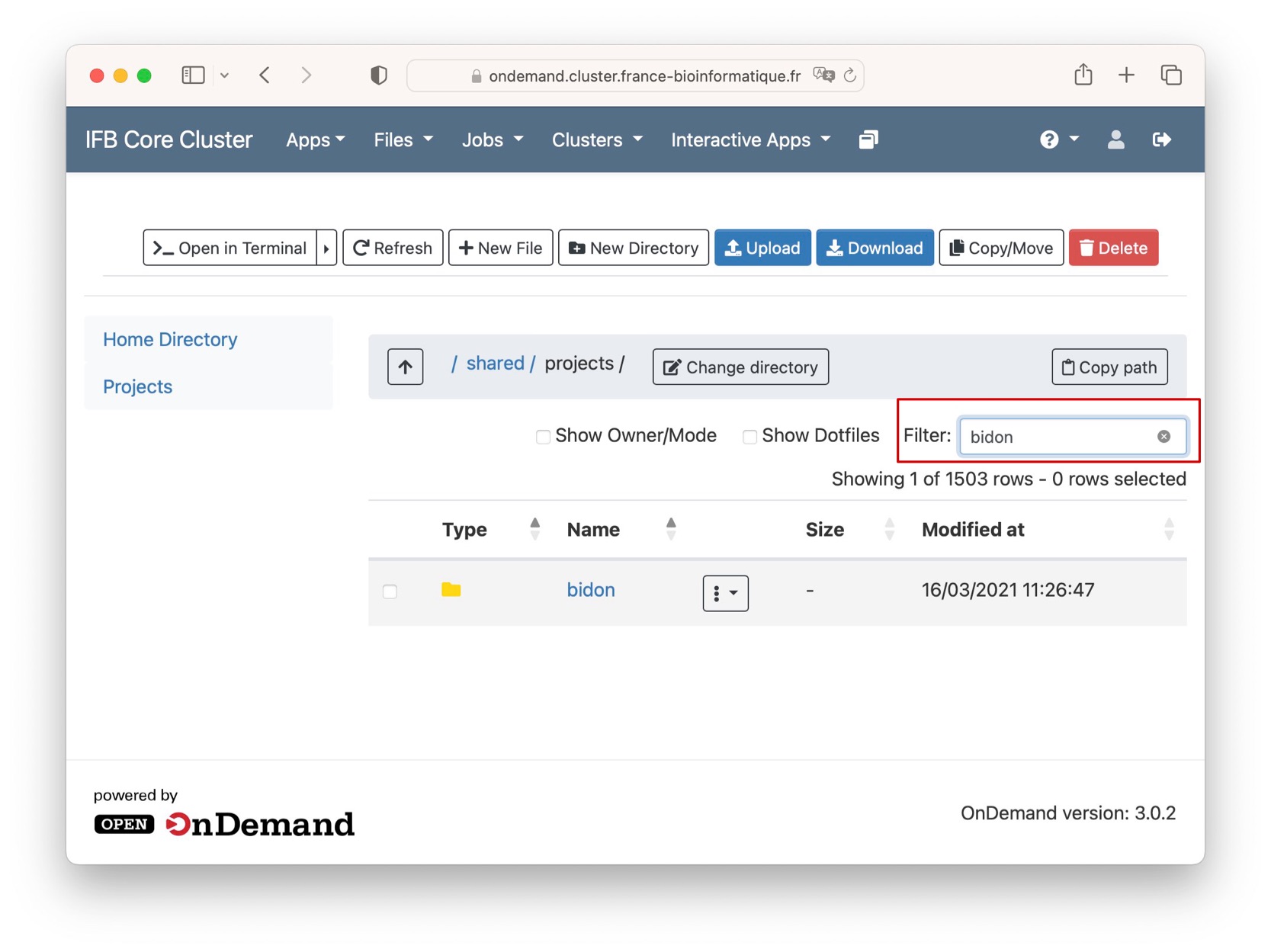
The projects space contains all the "projects" directories hosted on the infrastructure. Use the text box filter to find your project quickly.
Warning
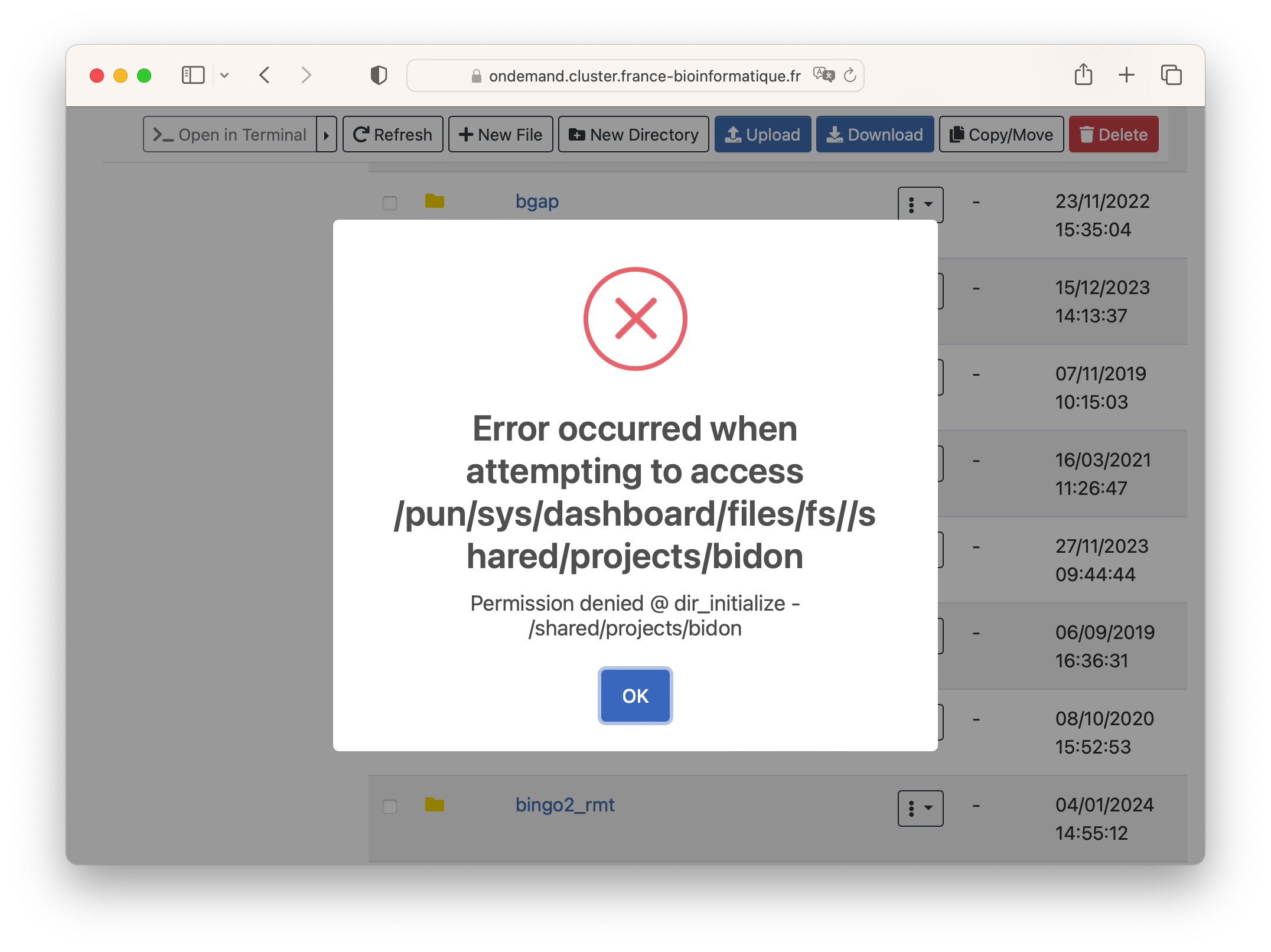
When you do not have access to a project, you will see an Error message like the one in the image below.
Jobs
Jobs can be monitored, created, edited and scheduled with the job management tools under the Jobs menu.
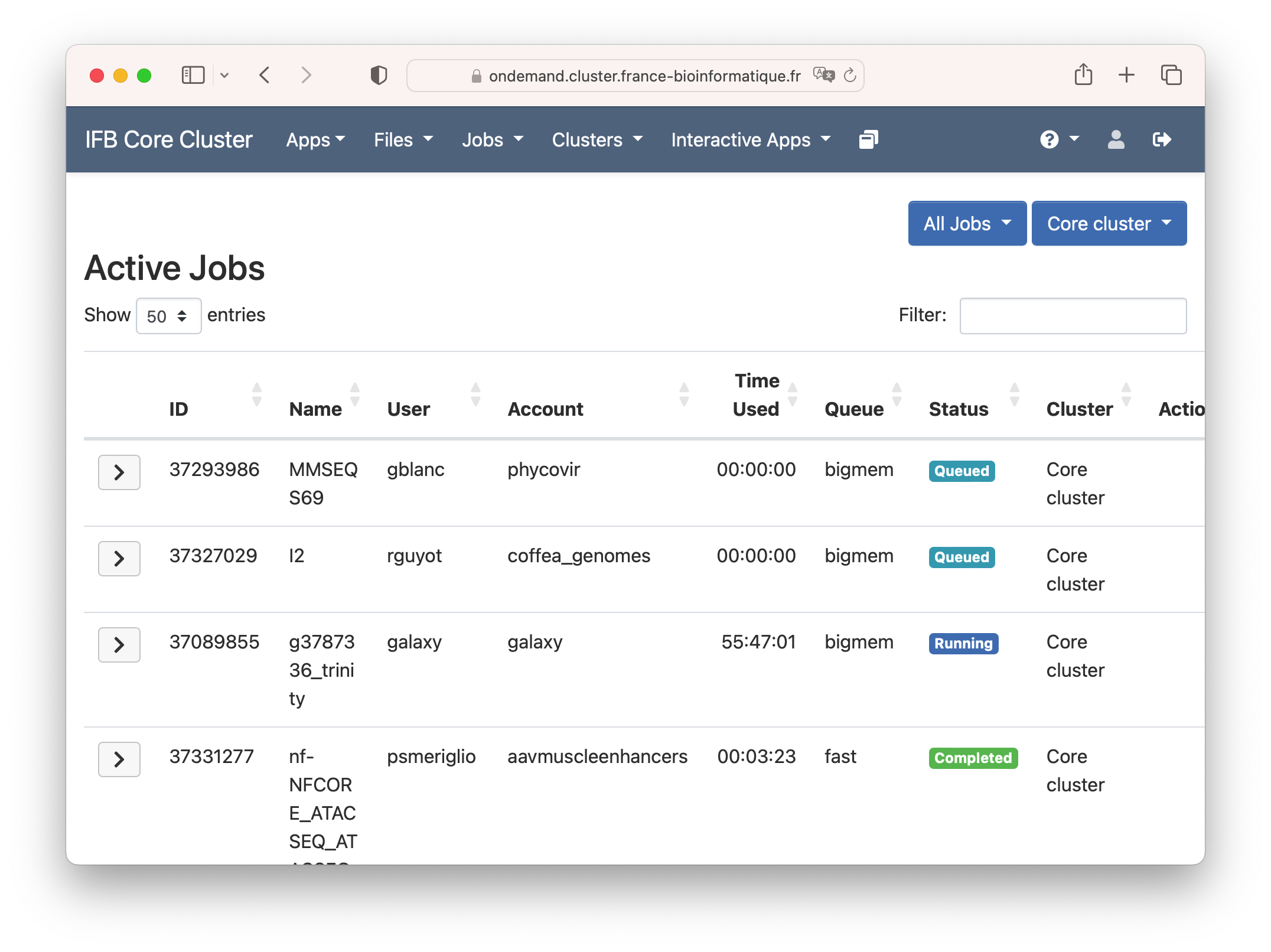
Under the "Active Jobs" tab you can view active jobs. You can choose to view your jobs or all jobs. From this menu you can also cancel your own jobs.
Create and edit job scripts and schedule jobs under this menu.
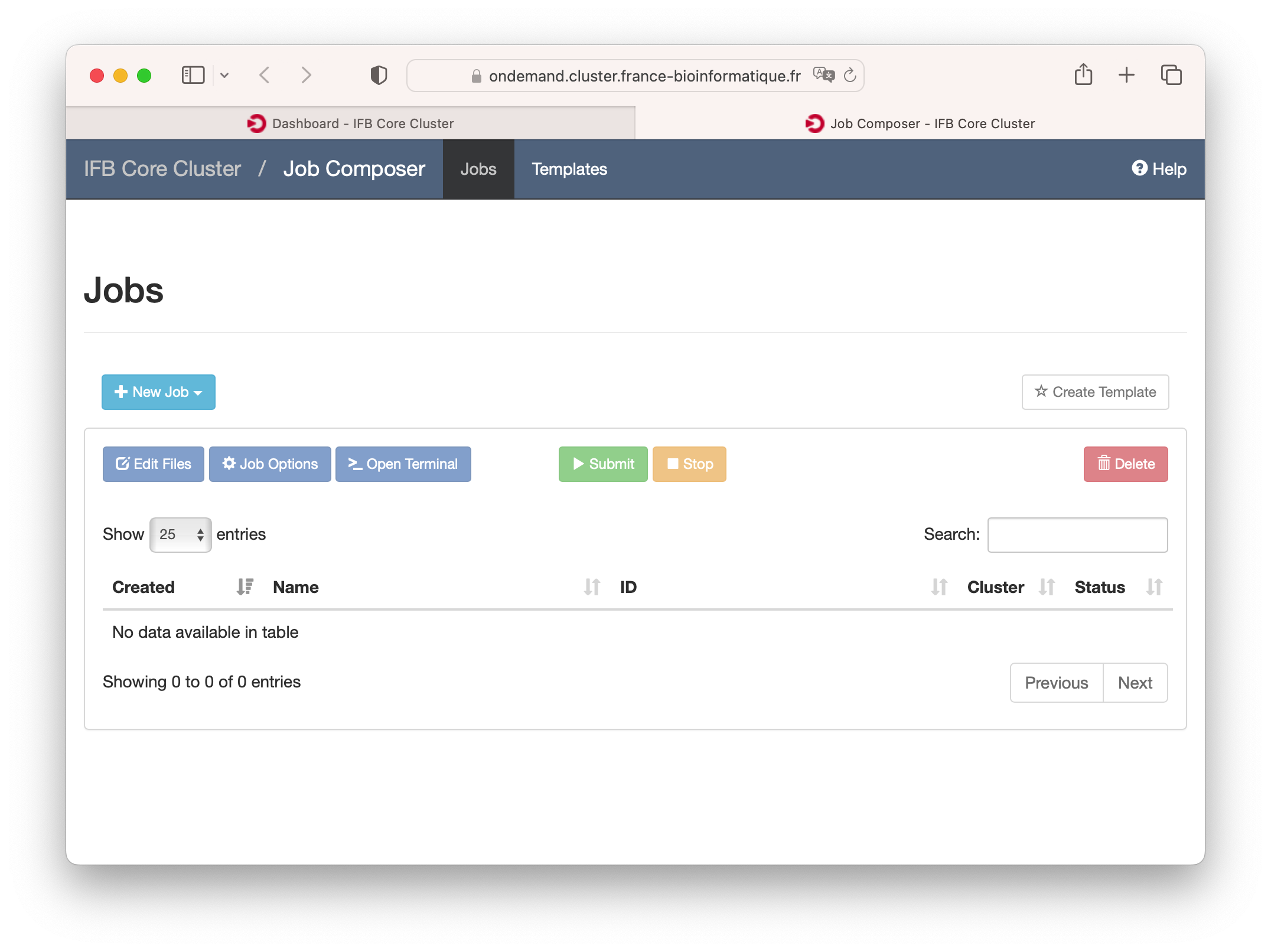
Clusters (Shell Access)
The Clusters menu offers shell access to the login node within the IFB cluster. This shell terminal operates similarly to various other tools providing terminal access.
Upon initiating the Terminal, you will be prompted to enter your IFB cluster account password. Once entered successfully (no characters will appear on the screen as you type), your shell session on the login node will begin.
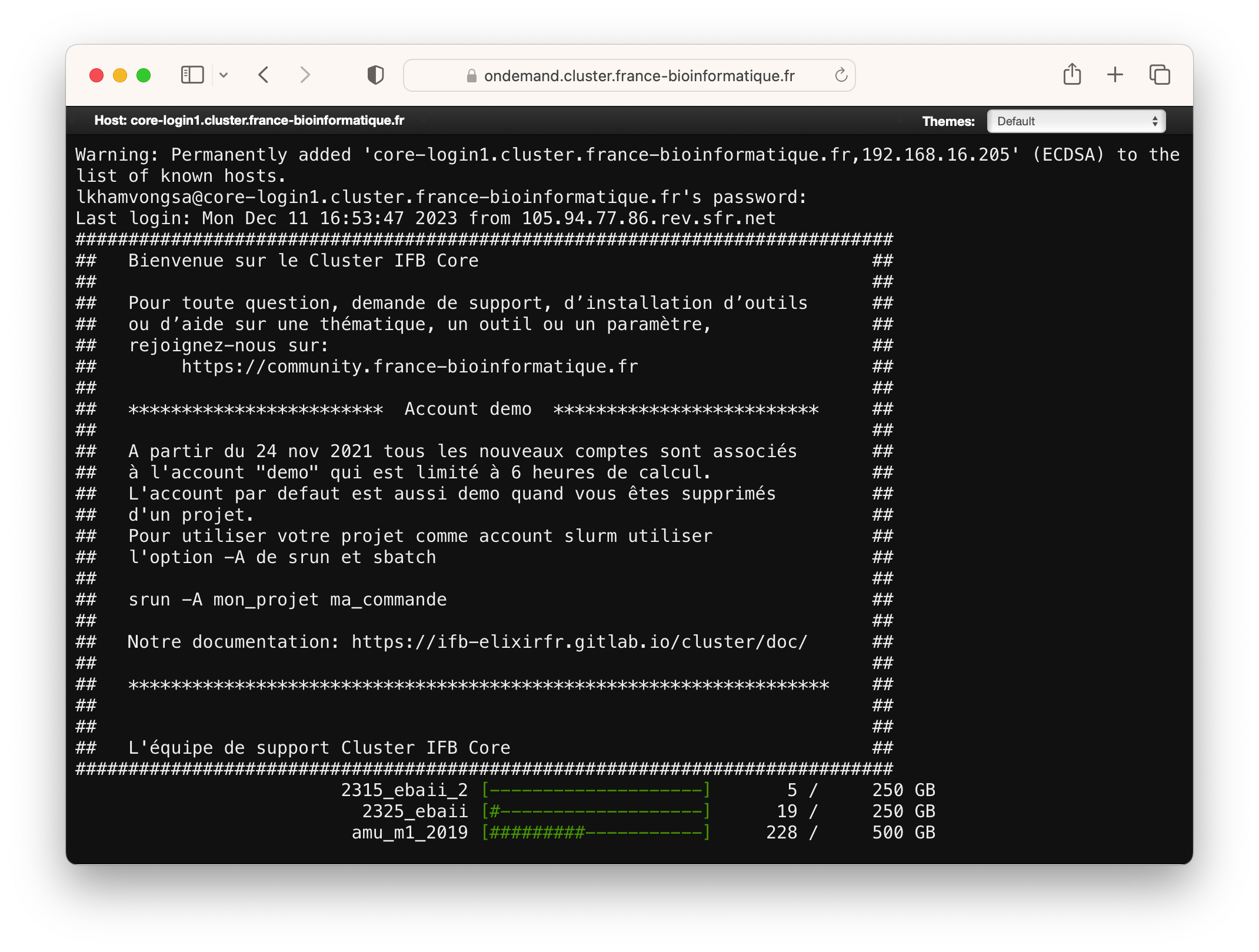
From here, you can work on the cluster using command line to run SLURM commands. If you are new to SLURM, please go through our documentations :
- SLURM at IFB Core
- SLURM user guide
- SLURM examples
- SLURM GPU
- SLURM advanced guide
- SLURM Cookbook
- SLURM Running interactive jobs
Warning
Please don't run any bioinformatics tools directly on the login node. The login node is not a compute node ; it has no resources to run heavy calculation.
Interactive Apps
The Interactive Apps menu contains options to launch certain applications that have graphical user interfaces (GUIs) for interactive use on IFB cluster. Each application is running in a dedicated SLURM jobs with the resources (RAM, CPU, GPU) you request.
Current supported applications include :
- JupyterLab : JupyterLab is a web-based interactive development environment for notebooks, code and data
- RStudio Server : RStudio Server is web based version of the RStudio integrated development envrionment (IDE) for R
To find out more about using apps, please consult the following documents :